Chapter 11
Monte Carlo Techniques
So far we have been focussing on how particle codes work once the
particles are launched. We've talked about how they are moved, and how
self-consistent forces on them are calculated. What we have not
addressed is how they are launched in an appropriate way in the first
place, and how particles are reinjected into a simulation. We've also
not explained how one decides statistically whether a collision has
taken place to any particle and how one would then decide what
scattering angle the collision corresponds to. All of this must be
determined in computational physics and engineering by the use of
random numbers and statistical distributions.66 Techniques based on random numbers
are called by the name of the famous casino at Monte Carlo.
11.1 Probability and Statistics
11.1.1 Probability and Probability Distribution
Probability, in the mathematically precise sense, is an
idealization of the repetition of a measurement, or a sample, or some
other test. The result in each individual case is supposed to be
unpredictable to some extent, but the repeated tests show some average
trends that it is the job of probability to represent. So, for
example, the single toss of a coin gives an unpredictable result:
heads or tails; but the repeated toss of a (fair) coin gives on
average equal numbers of heads and tails. Probability theory describes
that regularity by saying the probability of heads and tails is
equal. Generally the probability of a particular class of outcomes
(e.g. heads) is defined as the fraction of the outcomes, in a
very large number of tests, that are in the particular class. For a
fair coin toss, probability of heads is the fraction of outcomes of a
large number of tosses that is heads, 0.5. For a six-sided die, the
probability of getting any particular value, say 1, is the fraction of
rolls that come up 1, in a very large number of tests. That will be
one-sixth for a fair die. In all cases, because probability is defined
as a fraction, the sum of probabilities of all possible
outcomes must be unity.
More usually, in describing physical systems we deal with a
continuous real-valued outcome, such as the speed of a randomly
chosen particle. In that case the probability is described by a
"probability distribution" p(v)
which is a function of the random variable (in this case velocity
v). The probability of finding that the velocity lies in the range
v→ v+dv for small dv is then equal to p(v)dv. In order for the
sum of all possible probabilities to be unity, we require
Each individual sample67 might give rise to more than one
value. For example the velocity of a sampled particle might be a
three-dimensional vector v=(vx,vy,vz). In that case, the
probability distribution is a function in a multi-dimensional
parameter-space, and the probability of obtaining a sample that
happens to be in a multi-dimensional element d3v at v is
p(v)d3v. The corresponding normalization is
Obviously, what this shows is that if our sample consists of randomly
selecting particles from a velocity distribution function f(v), then
the corresponding probability function is simply
|
p(v) = f(v)/ | ⌠
⌡
|
f(v)d3v = f(v)/n, |
| (11.3) |
where n is the particle density. So the normalized distribution
function is the velocity probability distribution.
The cumulative probability
function can be considered to represent the probability that a sample
value is less than a particular value. So for a single-parameter
distribution p(v), the cumulative probability is
In multiple dimensions, the cumulative probability is a
multidimensional function that is the integral in all the dimensions
of the probability distribution:
|
P(v) = P(vx,vy,vz)= | ⌠
⌡
|
vx
−∞
|
| ⌠
⌡
|
vy
−∞
|
| ⌠
⌡
|
vz
−∞
|
p(v′)d3v′. |
| (11.5) |
Correspondingly the probability distribution is the derivative of the
cumulative probability: p(v)=dP/dv, or p(v)=∂3P/∂vx∂vy∂vz.
11.1.2 Mean, Variance, Standard Deviation, and Standard Error
If we make a large number N of individual measurements of a random
value from a probability distribution p(v), each of which gives a
value vi, i=1,2,...,N, then the
sample mean value of the combined sample N is defined as
The
sample variance is defined68 as
|
SN2 = |
1
N−1
|
|
N
∑
i=1
|
(vi−μN)2. |
| (11.7) |
The sample standard deviation
is SN, the square root of the
variance, and the
sample standard error is SN/√N. The mean is
obviously a measure of the average value, and the variance or standard
deviation is a measure of how spread out the random values are. They
are the simplest unbiassed estimates of the moments of the
distribution. These moments are properties of the
probability distribution not of the particular sample. The
distribution mean69 is defined as
and the distribution variance is
Obviously for large N we expect the sample mean to be approximately
equal to the distribution mean and the sample variance equal to the
distribution variance.
A finite size sample will not have a mean exactly equal to the
distribution mean because of statistical fluctuations. If we regard
the sample mean μN as itself being a random variable, which
changes from one total sample of N tests to the next total sample of
N tests, then it can be
shown70 that the probability distribution of
μN is approximately a Gaussian with standard deviation equal to
the standard error SN/√N. That is one reason why the
Gaussian distribution is
sometimes called the "Normal" distribution. The Gaussian probability
distribution in one dimension has only two71 independent
parameters μ and S.
11.2 Computational random selection
Computers can generate pseudo-random
numbers, usually by doing complicated non-linear arithmetic starting
from a particular "seed" number
(or strictly a seed "state"
which might be multiple numbers). Each successive
number produced is actually completely determined by the algorithm,
but the sequence has the appearance of randomness, in that the values
v jump around in the range 0 ≤ v ≤ 1, with no apparent
systematic trend to them. If the random number generator is a good
generator, then successive values will not have
statistically-detectable dependence on the prior values, and the
distribution of values in the range will be uniform, representing a
probability distribution p(v)=1. Many languages and mathematical
systems have library functions that return a random number. Not all
such functions are "good" random
number generators.
(The built-in C
functions are notoriously not good.) One should be wary for production
work. It is also extremely useful, for example for program debugging,
to be able to repeat a pseudo-random calculation, knowing the sequence
of "random" numbers you get each time will be the same. What you
must be careful about, though, is that if you want to improve the
accuracy of a calculation by increasing the number of samples, it is
essential that the samples be independent. Obviously, that means the
random numbers you use must not be the same ones you already
used. In other words, the seed must be different. This goes also for
parallel computations. Different parallel processors should normally
use different seeds.
Now obviously if our computational task calls for a random number from
a uniform distribution between 0 and 1, p(v)=1, then using one of
the internal or external library functions is the way to go. However,
usually we will be in a situation where the probability distribution
we want to draw from is non-uniform, for example a Gaussian
distribution, an exponential distribution, or some other function of value.
How do we do that?
We use two related random variables; call them u and v. Variable
u is going to be uniformly distributed between 0 and 1. (It is
called a "uniform deviate".) Variable v
is going to be related to u through some one-to-one functional relation.
Now if we take a particular sample value drawn from the uniform
deviate, u, there is a corresponding value v. What's more, we know
that the fraction of drawn values that are in a particular u-element du,
is equal to the fraction of values that are in the corresponding
v-element dv. Consequently, recognizing that those fractions are
pu(u)du and pv(v)dv respectively, where pu and pv are the
respective probability distributions of u and v, we have
|
pu(u)du=pv(v)dv ⇒ pv(v) = pu(u) | ⎢
⎢
|
du
dv
| ⎢
⎢
|
= | ⎢
⎢
|
du
dv
| ⎢
⎢
|
. |
| (11.10) |
The final equality uses the fact that pu=1 for a uniform deviate.
Therefore, if we are required to find random values v from a
probability distribution pv, we simply have to find a functional
relationship between v and u that satisfies pv(v) = |du/dv|. But we know of a function already that provides
this property. Consider the cumulative
probability Pv(v)=∫vpv(v′)dv′. It is monotonic, and ranges
between zero and 1. Therefore we may choose to write
|
u = Pv(v) for which |
du
dv
|
= pv(v). |
| (11.11) |
So if u=Pv(v), the v variable will be randomly distributed with
probability distribution pv(v). We are done. Actually not quite
done, because the process of choosing u and then finding the value
of v which corresponds to it requires us to invert the function
Pv(v). That is
Fig. 11.1 illustrates this process.
It is not always possible to invert the function analytically, but it
is always possible to do it numerically. One way is by root
finding e.g. bisection.
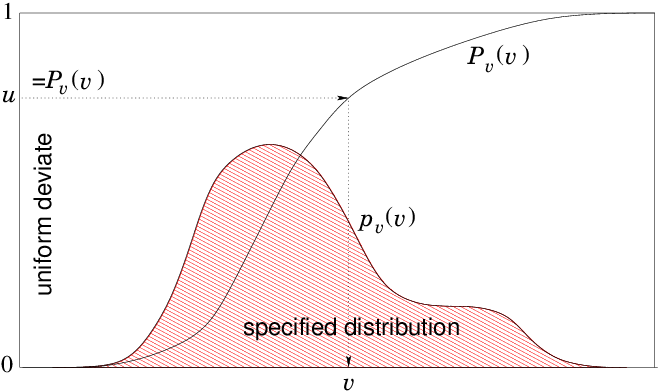
Figure 11.1: To obtain numerically a random variable v with specified
probability distribution p
v (not to scale), calculate a table
of the function P
v(v) by integration. Draw a random number from
uniform deviate u. Find the v for which P
v(v)=u by
interpolation. That's the random v.
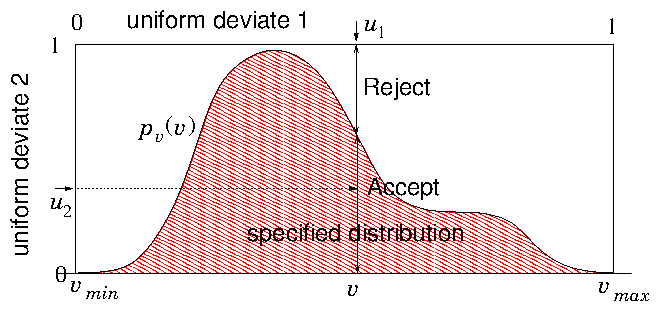
Figure 11.2: The rejection method chooses a v value randomly from a
simple distribution (e.g. a constant) whose integral is
invertible. Then a second random number decides whether it will
be rejected or accepted. The fraction accepted at v is equal
to the ratio of p
v(v) to the simple invertible distribution.
p
v(v) must be scaled by a constant factor to be everywhere
less than the simple distribution (1 here).
11.3 Flux integration and injection choice.
Suppose we are
simulating a sub-volume that is embedded in a larger region. Particles
move in and out of the sub-volume. Something interesting is being
modelled within the subvolume, for example the interaction of some
object with the particles. If the volume is big enough, the region
outside the subvolume is relatively unaffected by the physics in the
subvolume, then we know or can specify what the distribution function
of particles is in the outer region, at the volume's boundaries. Assume
that periodic boundary conditions are not appropriate, because, for
example, they don't well represent an isolated interaction. How do we
determine statistically what particles to inject into the subvolume
across its boundary?
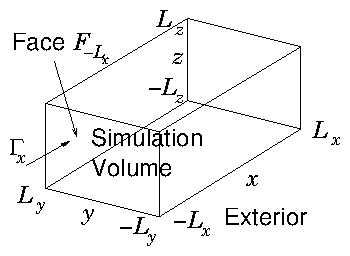
Figure 11.3: Simulating over a volume that is embedded in a wider
external region, we need to be able to decide how to inject
particles from the exterior into the simulation volume so as
to represent statistically the exterior distribution.
|
Γx(x) = | ⌠
⌡
|
| ⌠
⌡
|
| ⌠
⌡
|
∞
vx=0
|
vx f(v,x) dvx dvydvz |
| (11.13) |
and the number entering across the face per unit time (the flux) is
|
F−Lx = | ⌠
⌡
|
Ly
−Ly
|
| ⌠
⌡
|
Ly
−Ly
|
Γx(−Lx,y,z) dy dz. |
| (11.14) |
Assume that the outer region is steady, independent of time. We
proceed by evaluating the six fluxes Fj, using the above
expressions and their equivalents for the six different faces. At
each time step of the simulation, we decide how many particles to
inject into the subvolume from each face. The average number of
particles to inject in a timestep ∆t at face j is equal to
Fj∆t. If this number is large,74 then it may
be appropriate to inject just that number (although dealing with the
non-integer fractional part of the number needs consideration). But if
it is of order unity or even smaller, then that does not correctly
represent the statistics. In that case we need to decide statistically
at each step how many particles are injected: 0,1,2 ...
It is a standard result of probability theory 75 that
if events (in this case injections) take place randomly and
uncorrelated with each other at a fixed average rate r (per sample,
in this case per time-step) then the number n that happens in any
particular sample is an integer random variable with "Poisson
distribution": a discrete probability distribution
The parameter giving the rate, r, is a real number, but the number
for each sample, n, is an integer. One can rapidly verify that,
since
, the probabilities are
properly normalized:
. The mean rate is
(as advertized). The variance, it turns out, is also r. So the
standard deviation is √r. The value pn gives us precisely
the probability that n particles will need to be injected when the
flux is r=Fj. So the first step in deciding injections is to select
randomly a number to be injected, from the Poisson distribution eq. (11.15). There are various ways to do this (including
library routines). The root-finding approach is easily applied,
because the cumulative probability function Pu(u) can be considered
to consist of steps of height pn at the integers n (and constant
in between).
Next we need to decide where on the surface each injection is going to
take place. If the flux density is uniform, then we just pick randomly
a position corresponding to −Ly ≤ y ≤ Ly and −Lz ≤ z ≤ Lz. Non-uniform flux density, however, introduces another distribution
function inversion headache. It's more work, but straightforward.
Finally we need to select the actual velocity of the particle. Very
importantly, the probability distribution of this selection is
not just the velocity distribution function, or the velocity
distribution function restricted to positive vx. No, it it the
flux distribution vxf(v,x) weighted by the
normal velocity vx (for an x-surface) if positive (otherwise
zero). If the distribution is separable,
f(v)=fx(vx)fy(vy)fz(vz), as it is if it is Maxwellian,
then the tangential velocities vy, vz, can be treated
separately: select two different velocities from the respective
distributions. And select vx (positive) from a probability
distribution proportional to vxfx(vx).
If f is not separable, then a more elaborate random selection is
required. Suppose we have the cumulative probability distribution of
velocities P(vx,vy,vz) for all interesting values of
velocity. Notice that if vxmax, vymax, vzmax denote the
largest relevant values of vx, vy and vz, beyond which f=0,
then P(vx,vymax,vzmax) is a function of just one variable
vx, and is the cumulative probability distribution integrated over
the entire relevant range of the other velocity components. That is,
it is the one-dimensional cumulative probability distribution of
vx. We can convert it into a
one-dimensional
flux cumulative probability by performing the following
integral (numerically, discretely).
| |
|
|
| ⌠
⌡
|
vx
0
|
vx′ |
∂
∂ vx′
|
P(vx′,vymax,vzmax) dvx′ |
| | (11.16) |
| |
|
| vx P(vx,vymax,vzmax)− | ⌠
⌡
|
vx
0
|
P(vx′,vymax,vzmax) dvx′. |
| |
|
Afterwards, we can normalize F(vx) by dividing by F(vxmax),
ariving at the cumulative flux weighted probability for vx.
We then proceed as follows.
| 1. | Choose a random vx from its cumulative flux-weighted probability
F(vx). |
| 2. | Choose a random vy from its cumulative probability for the
already chosen vx, namely P(vx,vy,vzmax) regarded as a
function only of vy. |
| 3. | Choose a random vz from its cumulative probability for the
already chosen vx and vy, namely P(vx,vy,vz) regarded as a
function only of vz. |
Naturally for other faces, y and z one has to
start with the corresponding velocity component and cycle round the
indices. For steady external conditions all the cumulative velocity
probabilities need to be calculated only once, and then
stored for subsequent time steps.
Discrete particle representation An alternative to this
continuum approach is to suppose that the external distribution
function is represented by a perhaps large number, N, (maybe
millions) of representative "particles" distributed in accordance
with the external velocity distribution function. Particle k has
velocity vk and the density of particles in phase space is
proportional to the distribution function, that is to the probability
distribution. Then if we wished to select randomly a velocity from the
particle distribution we simply arrange the particles in order and
pick one of them at random. However, when we want the particles to be
flux-weighted, in normal direction
say, we must select
them with probability proportional to
(when
positive, and zero when negative). Therefore, for this normal
direction we must consider each particle to have appropriate weight.
We associate each particle with a real76 index r so that when rk ≤ r < rk+1 the particle k is indicated. The interval length
allocated to particle k is chosen proportional to its weight, so
that
. Then the
selection consists of a random number draw x, multiplied by the
total real index range and indexed to the particle and hence to its
velocity: x(rN+1−r1)+r1 = r → k → vk. The discreteness
of the particle distribution will generally not be an important
limitation for a process that already relies on random particle
representation. The position of injection will anyway be selected
differently even if a representative particle velocity is selected
more than once.
Worked example: High Dimensionality Integration
Monte Carlo techniques are commonly used for high-dimensional
problems; integration
is perhaps the simplest example. The
reasoning is approximately this. When there are d dimensions, the
total number of points in a grid whose size is M in each
coordinate-direction is N=Md. The fractional uncertainty in
estimating a one-dimensional integral of a function with only isolated
discontinuities, based upon evaluating it at M grid points, may be
estimated as ∝ 1/M. If this estimate still applies to multiple
dimension integration (and this is the dubious part), then the
fractional uncertainty is ∝ 1/M=N−1/d. By
comparison, the uncertainty in a Monte-Carlo estimate of the integral
based on N evaluations is ∝ N−1/2. When d is larger
than 2, the Monte Carlo square-root convergence scaling is better than
the grid estimate. And if d is very large, Monte Carlo is much
better. Is this argument correct? Test it by obtaining the volume of
a four-dimensional hypersphere by numerical
integration, comparing a grid technique with Monte Carlo.
A four-dimensional sphere of radius 1 consists of all those points for
which r2=x12+x22+x32+x42 ≤ 1. Its volume is known
analytically; it is π2/2. Let us evaluate the volume numerically
by examing the unit hypercube 0 ≤ xi ≤ 1, i=1,...,4. It is
1/24=1/16th of the hypercube −1 ≤ xi ≤ 1, inside of which the
hypersphere fits; so the volume of of the hypersphere that lies
within the 0 ≤ xi ≤ 1 unit hypercube is 1/16th of its total
volume; it is π2/32. We calculate this volume numerically by discrete
integration as follows.
A deterministic (non-random) integration of the volume consists of
constructing an equally spaced lattice of points at the
center of cells that fill the unit cube. If there are M points per
edge, then the lattice positions in the dimension i (i=1,...,4)
of the cell-centers are xi,ki=(ki−0.5)/M, where ki=1,...,M
is the (dimension-i) position index. We integrate the volume of the
sphere by collecting values from every lattice point throughout the
unit hypercube. A value is unity if the point lies within the
hypersphere r2 ≤ 1; otherwise it is zero. We sum all values (zero
or one) from every lattice point and obtain an integer equal to the
number of lattice points S inside the hypersphere. The total number
of lattice points is equal to M4. That sum corresponds to the total
volume of the hypercube, which is 1. Therefore the discrete estimate
of the volume of 1/16th of the hypersphere is S/M4. We can
compare this numerical integration with the analytic value and express
the fractional error as the numerical value divided by the analytic
value, minus one:
|
Fractional Error = | ⎢
⎢
|
S/M4
π2/32
|
−1 | ⎢
⎢
|
|
|
Monte Carlo integration works essentially exactly the same except that
the points we choose are not a regular lattice, but rather they are
random. Each one is found by taking four new uniform-variate values
(between 0 and 1) for the four coordinate values xi. The point
contributes unity if it has r2 ≤ 1 and zero otherwise. We obtain a
different count Sr. We'll choose to use a total number N of random point
positions exactly equal to the number of lattice points N=M4,
although we could have made N any integer we like. The Monte Carlo
integration estimate of the volume is Sr/N.
I wrote a computer code to carry out these simple procedures, and
compare the fractional errors for values of M ranging from 1 to
100. The results are shown in Fig. 11.4.
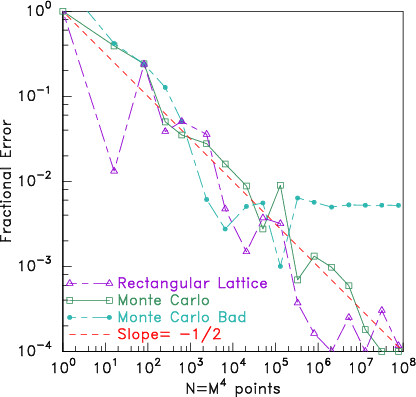
Figure 11.4: Comparing error in the volume of a hypersphere found
numerically using lattice and Monte Carlo integration. It turns out
that Monte Carlo integration actually does
not
converge significantly faster than lattice integration, contrary to
common wisdom. They both converge approximately like
1/√N (logarithmic slope =−
1/
2). What's more, if one
uses a "bad" random number generator (the Monte Carlo Bad
line) it is possible the random integration will cease
converging at some number, because it gives only a finite-length
of independent random numbers, which in this case is exhausted
at roughly a million.
Exercise 11. Monte Carlo Techniques
1. A random variable is required, distributed on the interval 0 ≤ x ≤ ∞ with probability distribution p(x)=kexp(−kx), with k
a constant. A library routine is available that returns a uniform
random variate y (i.e. with uniform probability 0 ≤ y ≤ 1). Give formulas and an algorithm to obtain the required randomly
distributed x value from the returned y value.
2. Particles that have a Maxwellian distribution
|
f(v) = n | ⎛
⎝
|
m
2πk T
| ⎞
⎠
|
3/2
|
exp | ⎛
⎝
|
− |
mv2
2kT
| ⎞
⎠
|
|
| (11.17) |
cross a boundary into a simulation region.
(a) Find the cumulative probability distribution P(vn) that ought
to be used to determine the velocity vn normal to the boundary, of
the injected particles.
(b) What is the total rate of injection per unit area that should be used?
(c) If the timestep duration is ∆t, and the total rate of
crossing a face is r such that r∆t = 1, what probability
distribution should be used to determine statistically the
actual integer number of particles injected at each step?
(d) What should be the probability of 0, 1, or 2 injections?
3. Write a code to perform a Monto Carlo integration of the area
under the curve y=(1−x2)0.3 for the interval −1 ≤ x ≤ 1. Experiment with different total numbers of sample points, and
determine the area accurate to 0.2%, and approximately
how many sample points are needed for that accuracy.



